Environmental determinants of antimicrobial resistance uncovered in Indian tribal communities
A new study applying a One Health perspective has revealed that among three of India’s most isolated tribal communities in South India, the battle against antimicrobial resistance (AMR) may begin at the local water source. Researchers from the Anthropological Survey of India; Sahid Afrid Mollick, Gin Khan Khual, and Pulamaghatta N. Venugopal in collaboration with the University of Hyderabad; Dr. Anwesh Maile, Project Scientist and Prof. Hampapathalu Adimurthy Nagarajaram, used shotgun metagenomic sequencing to profile the gut microbiomes of three Particularly Vulnerable Tribal Groups (PVTGs) in South India. The findings suggest that the source of unprotected drinking water acts as a key ecological driver of AMR in the gut.

The team studied 103 healthy adults from the Irula, Jenu Kuruba, and Kurumba communities across Tamil Nadu, Karnataka, and Kerala. Participants provided fecal samples and socio-demographic information, including details about water source (stream vs. tubewell) and residential context (rural vs. urban). This integration of lifestyle data with metagenomic analysis allowed the researchers to link environmental exposure with variations in gut resistome composition.
By sequencing the entire microbial DNA from stool samples, the study identified and quantified antimicrobial resistance genes (ARGs), providing new insights into how environmental and lifestyle factors shape the spread of resistance in vulnerable human populations.
Introduction
Antimicrobial resistance (AMR) represents one of the most critical public health challenges of the 21st century, threatening the efficacy of antibiotics, antivirals, antifungals, and antiparasitic agents that form the foundation of modern medicine. Although resistance is an inherent outcome of microbial evolution, its emergence and global spread have been intensified by human activities, particularly the misuse and overuse of antimicrobials across healthcare, agriculture, and veterinary sectors.
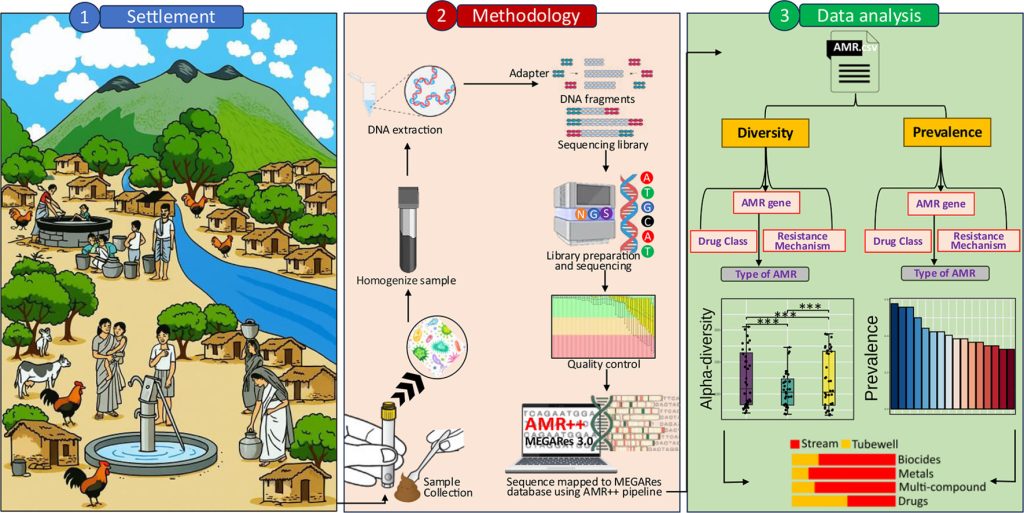
Visual representation that encapsulates the key components and findings of the study
Resistance emerged soon after antibiotics entered clinical practice. Penicillin resistance was documented as early as 1942, followed by tetracycline resistance in 1953 and the appearance of methicillin-resistant Staphylococcus aureus (MRSA) in the early 1960s. Subsequent decades saw the rise of multidrug-resistant tuberculosis (MDR-TB) and vancomycin-resistant pathogens, underscoring the escalating threat. Recognizing its magnitude, the World Health Organization (WHO) declared AMR a global health emergency in 2014.
Today, the world faces the looming risk of a “post-antibiotic era,” in which routine infections and minor injuries could once again become fatal. Without coordinated global action, AMR could cause up to 10 million deaths annually by 2050, posing serious implications for global health and development. In this context, the new study among India’s isolated tribal populations offers urgent insights into how local environments may shape the spread of resistance.
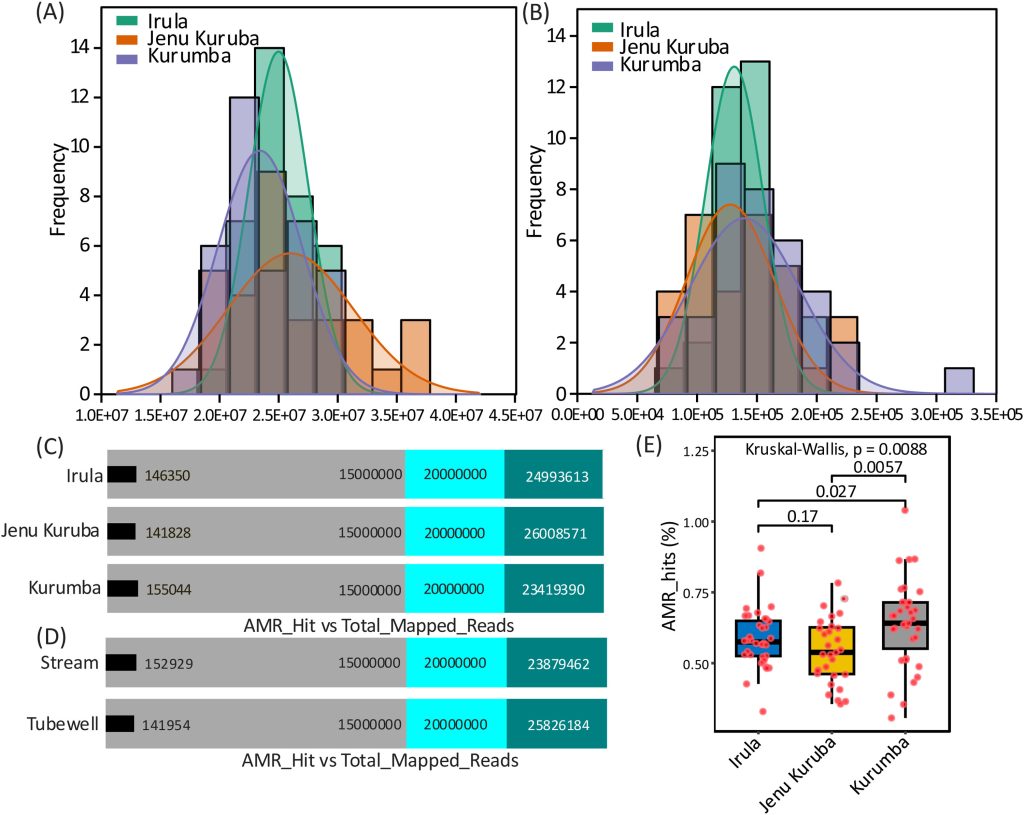
Distribution of processed reads and antimicrobial resistance (AMR) hit reads across the studied population.
A.Coverage distribution of total processed reads obtained from gut metagenomic sequencing.
B.Coverage distribution of reads mapped to AMR genes.
C.Comparison of total mapped reads and AMR-associated read counts across the population groups.
D.Comparison of total mapped reads and AMR-associated read counts stratified by drinking water habits.
E. Statistical evaluation of the percentage of AMR-associated reads among different population groups.
Limitation
This study’s cross-sectional design limits temporal assessment of gut resistome dynamics. While gut ARG profiles were correlated with drinking water sources, ARGs in water and other reservoirs (soil, food, livestock, or runoffs) were not examined. Moreover, only the genomic presence, not the functional activity or transfer potential, of AMR genes was assessed. Future studies should integrate environmental and functional analyses for a more comprehensive understanding of resistome transmission.
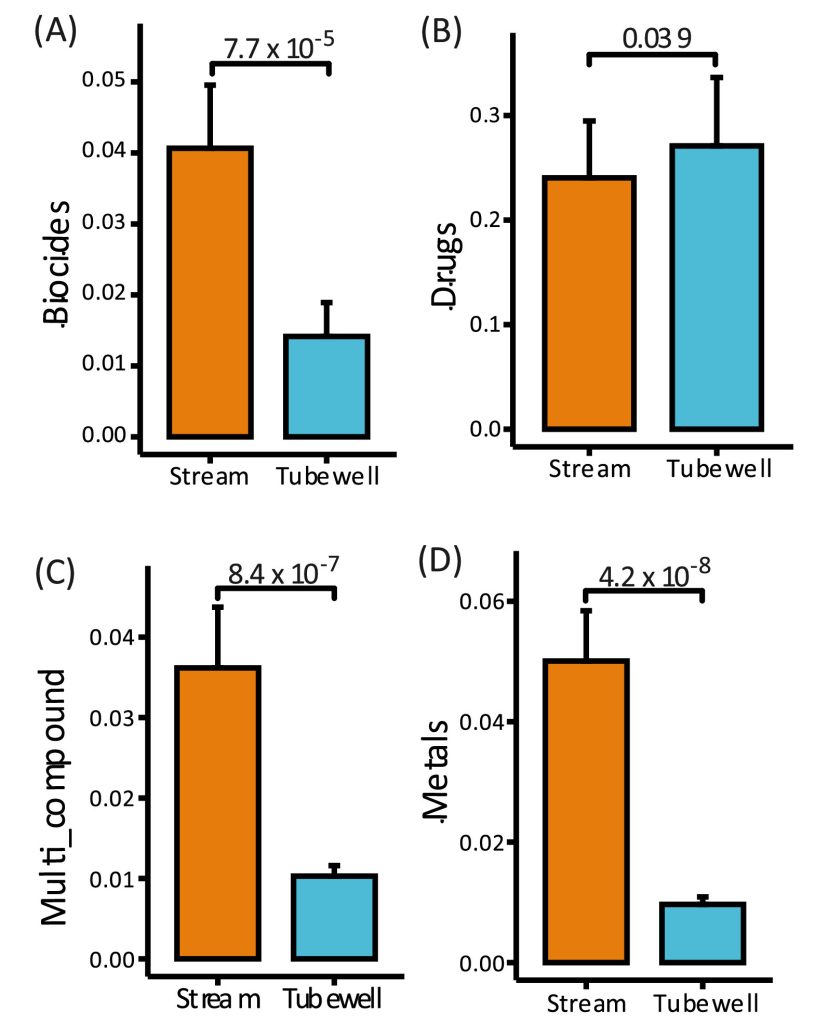
Distribution of Antimicrobial Resistance (AMR) Element Types in Relation to Drinking Water Source
This figure illustrates the distribution of AMR element types—biocides (A), antibiotics (B), multi-compound agents (C), and metals (D)—among individuals based on their primary drinking water source (stream vs. tubewell)
Conclusion
This study demonstrates that the drinking-water source is a key ecological driver of gut resistome variation in Indian tribal populations. While tubewell users predominantly harbor antibiotic resistance genes, stream-water consumers exhibit greater diversity and enrichment of non-antibiotic resistance determinants, particularly those linked to metals and biocides.
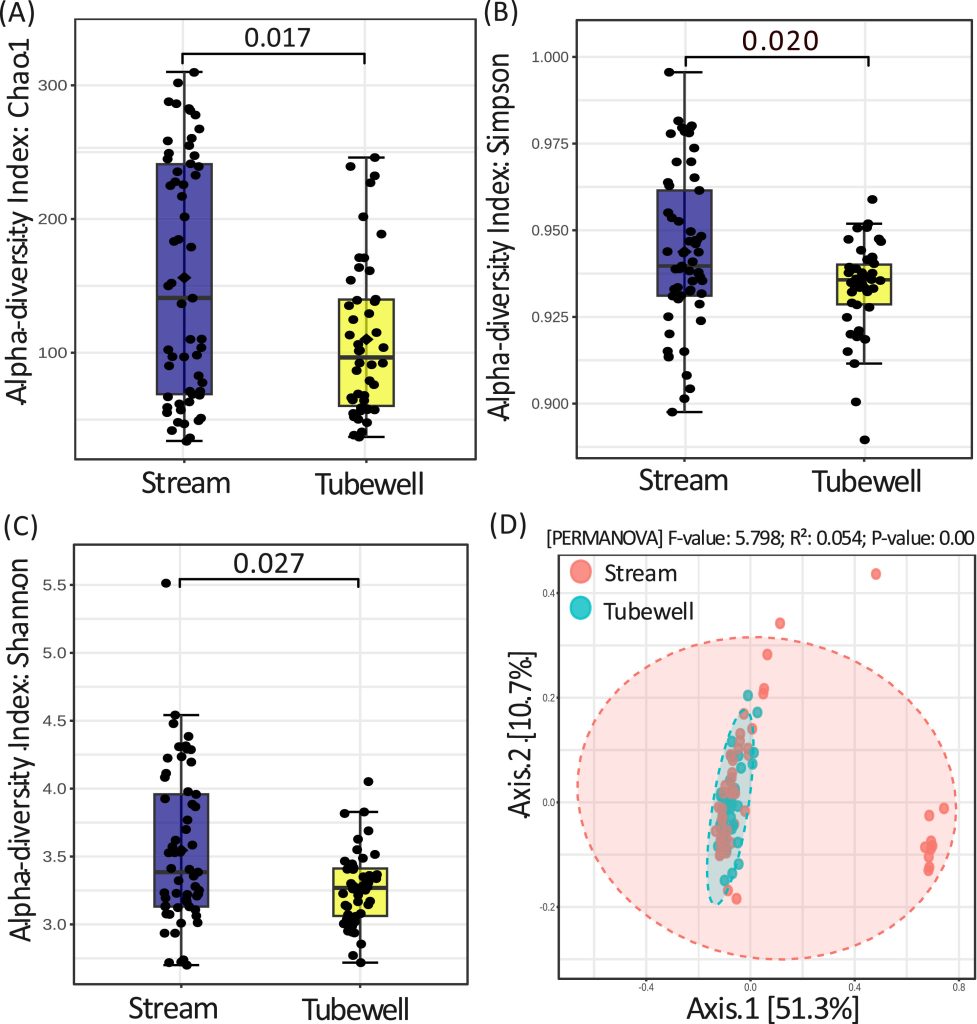
Compositional Diversity of Antimicrobial Resistance (AMR) Determinants in Relation to Lifestyle Factors with Emphasis on Drinking Water Source
A–C. Box plots showing intra-group diversity of AMR profiles among individuals consuming stream water versus tubewell water, assessed using multiple diversity indices.
D. Principal Coordinates Analysis (PCoA) illustrates inter-group variation in AMR composition and resistance mechanisms between stream and tubewell water consumers.
Highlights of the study:
- Source of drinking water shaped the resistome: individuals consuming stream water carried more metal, biocide, and multi-compound resistance, whereas tubewell water users had higher antibiotic resistance.
- Individuals consuming stream water showed significantly greater resistome diversity and ecological enrichment.
- Metal resistance genes were significantly overrepresented in microbial communities of individuals consuming stream water.
- Environmental co-selective pressures, particularly exposure to heavy metals, shaped resistome dynamics even with minimal antibiotic use.
- Findings highlight the need for safe water access, environmental surveillance, and resistome-informed public health strategies.
The details of the study can be read at: https://www.sciencedirect.
For any further information please email Prof. Nagarajaram HA at: hansl@uohyd.ac.in and can visit the site DBT- Centre for Microbial Informatics (DBT-CMI) (https://dbtcmi.in)